| |
If you have any questions about the resources
here, feel free to contact Kendrick.
TOOLS
We create a variety of software
tools and share these freely with the community.
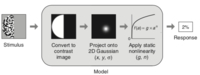 |
analyzePRF
This is a toolbox for estimating
population receptive fields from
time-series data (or from single-response
data). For more information, see the analyzePRF
web site and the analyzePRF
github repository. The
toolbox is closely related to the
compressive spatial summation Journal of
Neurophysiology paper.
|
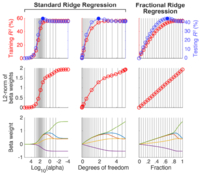 |
fracridge
This is a toolbox that implements
the fractional
ridge regression technique as
described in the GigaScience
paper. The primary advantage of
this implementation of ridge
regression is that you automatically get
correctly set hyperparameters
that probe the
relevant range for any regression
problem. For
more information, see the fracridge
github repository.
|
 |
GLMdenoise
This is a toolbox for denoising
task-based fMRI data. The basic
principle is to derive nuisance
regressors in a data-driven fashion
and to use cross-validation to
determine the optimal number of regressors (to avoid overfitting).
It was
originally introduced in this
Frontiers
paper and followed up in this
NeuroImage
paper.
For more information, see the GLMdenoise
web site and the GLMdenoise
github repository. (GLMsingle is a
replacement for GLMdenoise; see below.)
|
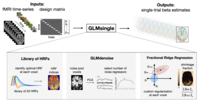 |
GLMsingle
This toolbox provides new methods for
improved single-trial GLM estimation in
fMRI time-series data. GLMsingle can be
viewed as a wholesale replacement of
GLMdenoise. GLMsingle incorporates (1) HRF
estimation per voxel (using a
library-of-HRFs approach), (2) data-driven
denoising (using GLMdenoise), and (3)
regularized single-trial estimation using
fractional ridge regression. The technique
was introduced in the NSD
data paper and is given a fuller
treatment in this
pre-print. For more
information, see glmsingle.org.
|
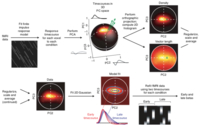 |
TDM
This is a toolbox that implements the Temporal Decomposition
Method as
described in the
Nature Methods paper. This TDM
technique provides a
method for summarizing, visualizing,
and extracting different response timecourses
from fMRI data. These timecourses
can then be used to isolate early and late
components of fMRI responses
associated with microvasculature
and macrovasculature,
respectively. For
more information, see the TDM
github repository.
|
IN-HOUSE TOOLS
These tools are primarily
designed and intended for in-house projects, but
might be useful more generally (depending on one's
workflow).
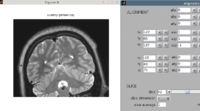 |
alignvolumedata
This toolbox provides
a convenient visual interface
for achieving
manual or automatic co-registration of
two 3D volumes. Rigid-body
and affine transformations are allowable. See the alignvolumedata
github repository.
|
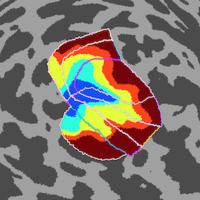 |
cvncode
This
collection of utilities handles
various pre-processing procedures
related to fMRI analysis. The most useful
functions are ones that
pertain to automatic
generation of visualizations
of surface-based data (cvnlookup) and
to
manual
drawing of ROIs
on
surface
data
(cvndefinerois).
See
the cvncode
github
repository.
|
 |
knkutils
Over
the years, we have collected a large
number of utility functions
that facilitate computational
analyses.
Some of these utilities are
incorporated into other of
our repositories, but in some cases, the knkutils repository is a
required dependency. See the knkutils github
repository.
|
 |
preprocessfmri
This toolbox implements
volume- and surface-based
pre-processing of fMRI data, including
dynamic time-varying
fieldmap-based
corrections and generation of
extensive visual inspections. Having been
developed over many years, it is
flexible,
powerful, but a bit
complex. See
the preprocessfmri
github repository.
|
DATASETS
We share data and code from our
fMRI experiments.
 |
Natural
Scenes Dataset
This
dataset consists of
massive dense sampling of whole-brain
7T fMRI responses to
color natural scenes in eight
carefully screened observers. We
also provide an archive of code used in this project.
For more
information, see the NSD web site
and the nsddatapaper
github repository.
|
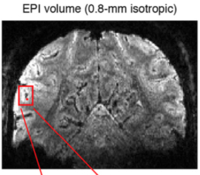 |
Ultra-high-resolution
fMRI in visual cortex
We
collected a number of high-resolution
(0.8-mm) 7T fMRI datasets using basic visual
manipulations. The
data formed the basis of
several papers: the veins
NeuroImage paper,
the TDM
technique Nature
Methods paper, and
a ventral visual
cortex Journal of
Neuroscience paper.
For more information,
see this
OSF
web site.
|
 |
HCP7T Retinotopy Dataset
A
large collection
(n = 181) of
fMRI retinotopic mapping data collected
as part of the Human Connectome Project.
The dataset is formally described in
the HCP7TRET
Journal of Vision paper. For
more information, see the HCP7TRET
OSF site.
|
 |
vtcipsmodel
This
dataset is associated with the bottom-up/top-down eLife paper. The experiment involved
a wide range of face and word
stimuli under different stimulus
manipulations and task
manipulations. We also include the
code we used to
model ventral visual regions and
their interaction with parietal
cortex. For more
information, see the vtcipsmodel
github repository.
|
 |
vtcdata
This
dataset is associated with
the face
pRF Current Biology paper. The
experiment involved
mapping population receptive
fields in face-selective regions
of human visual cortex under
different behavioral states. For
more information, see the vtcdata
web site.
|
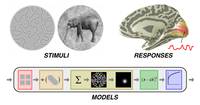 |
socmodel
This
dataset is associated with the second-order contrast (SOC)
PLoS Computational Biology paper.
The dataset includes fMRI responses in
visual cortex to a wide range of
synthetic stimuli, and also
includes code implementing the
SOC model described in the paper.
For more information, see
the socmodel
web site.
|
 |
vim-1
This
dataset is associated with the encoding/decoding
Nature paper,
and consists of
fMRI responses in occipital cortex
to a large number of grayscale natural
images. The data are hosted at crcns.org.
|
TEACHING
 |
The
Big Data Revolution in Neuroscience
A
week-long workshop held at NYU Abu Dhabi
in January 2020. Materials
include lecture
slides, videos, and code. The
content largely
revolves around the design,
pre-processing, and analyses that can be performed
on the very large Natural
Scenes Dataset.
|
 |
Statistics and Data
Analysis in MATLAB
A
course that we developed
to cover fundamental
principles in statistics as well as
how to implement and deal with these concepts effectively in code.
Materials include lecture
slides,
videos, and code. Be sure to
check out the figures and animations
that illustrate core statistical
concepts.
|
 |
Blog
on statistical analyses
A series of short
blog posts on some interesting observations
in computational
statistics.
|
 |
Cognitive
neuroscience links
A
collection of links
to demos and materials available
on the web that help
illustrate concepts in cognitive
neuroscience.
|
SEMINARS / TALKS
 |
Recorded talks
MRC-CBU
Chaucer Club 2021 talk on "A
data-driven approach to advancing
cognitive neuroscience" (video)
Algonauts workshop 2019 talk on the
NSD dataset (video
| slides)
OSA
Fall Vision Meeting 2017 talk on the
bottom-up/top-down eLife paper (video
| slides)
BrainHack
2017 talk on fMRI software
tools
(video
| slides)
Neurohackweek
2016
talk on modeling fMRI
data (video
| slides)
VSS
2016 symposium
talk on "what are
deep neural
networks and what
are they good
for?" (video
| slides)
VSS
2014 symposium
on "understanding
representation
in visual
cortex: why
are there so
many
approaches and
which is
best?" (videos)
|
OTHER
 |
Brain
art
In
several MRI
projects (especially those pitched
at ultra-high spatial resolution fMRI),
careful and informative
visualizations play a key role (see
the veins
NeuroImage paper). Some of these explorations
have generated some visually interesting figures, we
have compiled these
into a Google
Photos album.
|
|
|
|
|





















